By Vince Giuliano
Recent research indicates a brighter possibility for extraordinary human longevity – healthy lives extending perhaps to hundreds of years. The general approach is one I have been discussing for years, Closing the loop in the stem cell supply chain. Important recent specific progress was brought to my attention by Dr. Victoria Lunyak, a lab director at the Buck Institute who presented on her recent work at the Cell signaling, inflammation and aging symposium in Las Vegas last week, where I was also a presenter. This blog describes key recent work of Dr. Lunyak and others that lends potential reality to the vision of extraordinary human longevity.
Image: The six symbols for longevity
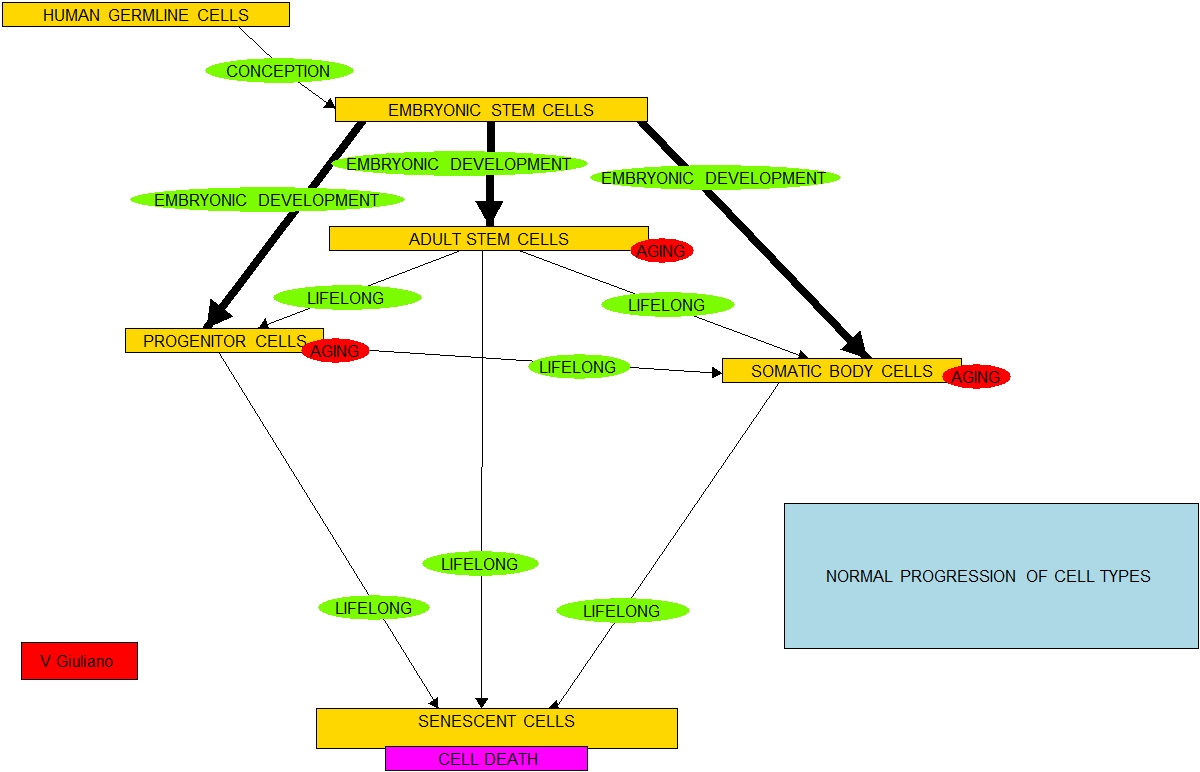
About adult stem cells: relationship to health and longevity and senescence
Life depends on constant replenishment of human body cells with new cells created by differentiation of adult stem cells. Many human body cell types have a remarkable rate of turnover. For example, under normal conditions, red blood cells (RBCs) have a lifespan of about 120 days and from one to five million new red blood cells are produced per second(ref). As Dr.Lunyak pointed out in her presentation, 50-70 million cells are discarded and new ones are generated daily. “In humans, it is estimated that keratinocytes turnover from stem cells to desquamation (skin peeling) every 40–56 days[7] whereas in mice the estimated turnover time is 8–10 days[8](ref).” Health depends critically on the replacement process.
As I originally described the stem cell supply chain:
“According to a simplified model of this theory a newly-conceived human embryo consists of pluripotent stem cells (Type A), ones that can potentially divide into any body cells. With growth, these proliferate and, in a remarkably articulated manner, progressively differentiate into multipotent stem cells (Type B), progenitor cells (Type C), mature body somatic cells (Type E), and many eventually become senescent cells (Type E).
According to the best current understanding of stem cells this is an open-loop process. The above list is in order of increasing cell-type specificity and decreasing cell-type potency to differentiate into other cell types. Starting at conception and throughout life, all cells on this list except the senescent ones will selectively reproduce and possibly differentiate into cells of types further down in the list. The state of the body in terms of makeup of cell types continues to change through life and the process goes inexplicably from start (conception) leading to end (death).
At conception, the embryo is all Type A cells. At maturity there are relatively very few Type A cells and a mix of Type B, C and D cells, Type B and C cells typically live in protected stem cell niches where they reproduce and, as-needed differentiate to become the normal working body Type D cells. As Type D cells die from trauma or apoptosis they are replaced by new cells resulting from differentiation of Type B and Type C cells. Stem cell gene expression evolves with age. “In newborn mice, blood-forming cells (hematopoietic stem cells, HSCs) rely on a transcription factor known as Sox17 for self-renewal, but adult HSCs rely on a different transcription factor, Bmi-1(ref).” At an advanced age, the pools of Type B and Type C cells become depleted in part because of replicative senescence and the cells remaining in the pools lose their ability to differentiate as necessary to replace Type D cells.
Although in principle stem cells can replicate indefinitely, in fact they age as the organism ages, continuing to change their gene expression. And the gene expression changes in a way that favors protection against cancer over differentiation capability, e.g. expression of p16ink4a increases. Many Type D cells senesce and become Type E cells which make the corresponding organs shrivel and be susceptible to cancers and other disease processes.
In essence, early-on the body sets up pools of stem and progenitor cells to replace lost somatic cells. Cells in those pools replicate and differentiate throughout life. But when these pools become compromised or depleted or the cells in them lose their capability to differentiate – well – to be blunt soon its curtains.”
That is the essence of the Stem Cell Supply Chain Breakdown theory of aging. IF we could find a way to rejuvenate adult stem cells in their niches, then the stem cell supply chain could possibly be transformed from being a once-through-in-life process to a continuing closed-loop process. And then, human life could possibly go on for a very long time.
This view appears to be consistent with that of some but not all researchers. For example, see Adult stem cells: simply a tool for regenerative medicine or an additional piece in the puzzle of human aging? (Tollervey JR, Lunyak V), Dec. 2011, and also some of the recent technical publications cited below.
Stem cell senescence is an epigenetic phenomenon.
Oxidative stress may well be a factor contributing to hASC senescence, and age-related shifts in niche signaling like Notch is also clearly involved(ref). However, I think the fundamental cause of senescence is epigenetic shifts leading to progressive gene silencing. Senescent cells tend to have shorter telomeres, but I believe this is mainly a downstream and not causal effect.
The 2008 review publication Epigenetic regulation of stem cell fate (Lunyak VV, Rosenfeld MG.) outlines the role of epigenetic factors in adult stem cell senescence and aging. “The multipotency of stem cells is reduced over time due to progressive gene silencing. Genes active in earlier progenitors are gradually silenced at developmentally later stages, and subsets of cell type-specific genes are turned on. This progression is the result of selective expression of transcription factors (TFs) in concert with classis ‘corner stones’ of epigenetics – chromatin remodeling and chromatin modifications, DNA methylation of CpG dinucleotides (4,5). As a result of these events compactization of the chromatin, its accessibility and positioning within specialized nuclear domains undergo dynamic changes. For example, it has been shown that heterochromatic markers, such as HP1 proteins, as well as heterochromatic histone modifications change their localization from dispersed and very dynamic in ESC to more concentrated distinct loci during cellular differentiation (6,7). This suggests that differentiation leads to the restructuring of the chromatin accompanied by the change in the global nuclear architecture, thus allowing the pluripotent nature of ESC genome to become more condensed, and, therefore, more transcriptionally restrained with maturation of the heterochromatin.” – “In mammalian cells, both DNA methylation and specific histone modification are involved in chromatin silencing. DNA methylation and histone modification are believed to be interdependent processes. Recent studies suggest that a combination of histone acetylation and DNA demethylation induces neuronal stem cells (NSC) to trans-differentiate into hematopoietic cells (8).”
Stem cell senescence results from aging and demonstrably leads to diseases and aging.
From the 2011 publication Manifestations and mechanisms of stem cell aging (Liu1,2 and Rando): “Aging is accompanied by a decline in the homeostatic and regenerative capacity of all tissues and organs (Kirkwood, 2005; Rando, 2006). With age, wound healing is slower in the skin, hair turns gray or is lost, skeletal muscle mass and strength decrease, the ratio of cellular constituents in the blood is skewed, and there is a decline in neurogenesis (Sharpless and DePinho, 2007). As the homeostatic and regenerative activities of these tissues are attributable to the resident stem cells, these age-related changes are reflections of declines in stem cell function (Bell and Van Zant, 2004; Dorshkind et. al, 2009; Jones and Rando, 2011). Clearly, in terms of organismal aging, the focus on stem cells is most relevant for those tissues in which normal cellular turnover is very high, such as epithelia of the skin and gut, as opposed to tissues, such as the cerebral cortex and the heart, in which cellular turnover in adults is exceedingly low (Rando, 2006). There is also an increasing interest in the therapeutic potential of stem cells to treat age-related degenerative diseases or conditions, further highlighting the importance of understanding the relationship between stem cell function and the properties of aged tissues.”
Epigenetic interventions can reverse cell senescence markers affecting aging.
According to the publication Histone Demethylase UTX-1 Regulates C. elegans Life Span by Targeting the Insulin/IGF-1 Signaling Pathway (Jin et. al.), 2011, “Like stem cell reprogramming, our results suggest that reestablishment of epigenetic marks lost during aging might help reset the developmental age of animal cells.” Regarding that publication, Lunyak and Kennedy had to say in Aged Worms Erase Epigenetic History: “Defining the molecular events that precipitate multisystem decline is an important component of aging research. In this issue, Jin et al., 2011 show that increased expression of the histone demethylase, utx-1, causes genome-wide decreases in histone H327 trimethylation, which includes the insulin/IGF-1 signaling (IIS) pathway that promotes aging.”
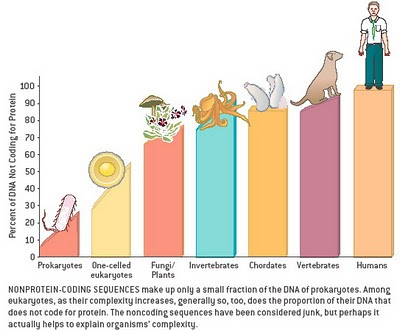
The genome goes far beyond DNA; it includes RNA and all so-called “junk” and non-coding sequences.
As a reminder for us that tend to think only in terms of DNA and genes: “In modern molecular biology and genetics, the genome is the entirety of an organism’s hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA.[1] (ref)”
New categories of both genomic and epigenomic knowledge are emerging, upsetting or refining ideas of just a few years ago.
1. One example is a re-evaluation of the roles of what was once thought to be “junk DNA,” often repeated and transposable segments of DNA not in genes.
The 2011 publication Genomicrelationship between SINE retrotransposons, Pol III-Pol II transcription, and chromatin organization: the journey from junk to jewel (Lunyak VV, Atallah M.) reports: “A typical eukaryotic genome harbors a rich variety of repetitive elements. The most abundant are retrotransposons, mobile retroelements that utilize reverse transcriptase and an RNA intermediate to relocate to a new location within the cellular genomes. A vast majority of the repetitive mammalian genome content has originated from the retrotransposition of SINE (100-300 bp short interspersed nuclear elements that are derived from the structural 7SL RNA or tRNA), LINE (7kb long interspersed nuclear element), and LTR (2-3 kb long terminal repeats) transposable element superfamilies. Broadly labeled as “evolutionary junkyard” or “fossils”, this enigmatic “dark matter” of the genome possesses many yet to be discovered properties.” As described below, such transposable segments may harbor the key to adult stem cell rejuvenation.
Image source
As a matter of fact, human retrotransposons represent for about 13% of the genome and account for about 50% of genetic information, around a million copies (Lunyak PowerPoint presentation). A transposon or “transposable element (TE) is a DNA sequence that can change its relative position (self-transpose) within the genome of a single cell(ref).”
2. Small non-coding RNA species (like IncRNAs, shRNAs, siRNAs and piALU RNAs) can play critical roles in gene regulation, DNA repair and chromatin regulation.
The 2012 publication Protein interactions with piALU RNA indicates putative participation of retroRNA in the cell cycle, DNA repair and chromatin assembly (Blackwell, Luynac et. al.) reports “Recent analyses suggest that transposable element-derived transcripts are processed to yield a variety of small RNA species that play critical functional roles in gene regulation and chromatin organization as well as genome stability and maintenance. Here we report a mass spectrometry analysis of an RNA-affinity complex isolation using a piRNA homologous sequence derived from Alu retrotransposal RNA. Our data point to potential roles for piALU RNAs in DNA repair, cell cycle and chromatin regulations.”
3. In fact, SINE/Alu Retrotransposons are transcriptionally up-regulated upon Senescence of hADSC, and this affects chromatin structure and impairs the DNA damage repair machinery.
(PowerPoint presentation by V. Lunyak). Also the same 2012 publication Protein interactions with piALU RNA indicates putative participation of retroRNA in the cell cycle, DNA repair and chromatin assembly (Blackwell, Lunyak et. al). With hADSC senescence, possibly associated with DNA methylation and age-related chromatin restructuring, there is a loss of regulation controlling SINE/ALU transcription and processing complexes.
Good news covered below is that by knocking down SINE/ALU transcription, hASC senescence can be reversed.
4. Another example is the emerging importance of intragenic DNA methylation, methylation within genes not at promoter sites.
For example, the 2012 publication Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research(Shenker N, Flanagan JM) reports: “Epigenetics is the study of all mechanisms that regulate gene transcription and genome stability that are maintained throughout the cell division, but do not include the DNA sequence itself. The best-studied epigenetic mechanism to date is DNA methylation, where methyl groups are added to the cytosine base within cytosine-guanine dinucleotides (CpG sites). CpGs are frequently clustered in high density (CpG islands (CGIs)) at the promoter of over half of all genes. Current knowledge of transcriptional regulation by DNA methylation centres on its role at the promoter where unmethylated CGIs are present at most actively transcribed genes, whereas hypermethylation of the promoter results in gene repression. – Over the last 5 years, research has gradually incorporated a broader understanding that methylation patterns across the gene (so-called intragenic or gene body methylation) may have a role in transcriptional regulation and efficiency. Numerous genome-wide DNA methylation profiling studies now support this notion, although whether DNA methylation patterns are a cause or consequence of other regulatory mechanisms is not yet clear. This review will examine the evidence for the function of intragenic methylation in gene transcription, and discuss the significance of this in carcinogenesis and for the future use of therapies targeted against DNA methylation.”
Further, the 2012 publication On the presence and role of human gene-body DNA methylation (Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK) reports: “DNA methylation of promoter sequences is a repressive epigenetic mark that down-regulates gene expression. However, DNA methylation is more prevalent within gene-bodies than seen for promoters, and gene-body methylation has been observed to be positively correlated with gene expression levels. This paradox remains unexplained, and accordingly the role of DNA methylation in gene-bodies is poorly understood. We addressed the presence and role of human gene-body DNA methylation using a meta-analysis of human genome-wide methylation, expression and chromatin data sets.”
5. Messenger RNA is subject to adenine methylation and a fifth nucleobases, N6-methyladenosine (m6A), is very common in the transcriptome. Up to 20 percent of human mRNA is routinely methylated. Since more than 5,000 different mRNA molecules contain m6A, mRNA methylation is likely to have widespread effects on how genes are expressed.
The 2012 publication Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3 UTRs and near Stop Codons (Meyer et. al.) reports “Methylation of the N6 position of adenosine (m6A) is a posttranscriptional modification of RNA with poorly understood prevalence and physiological relevance. The recent discovery that FTO, an obesity risk gene, encodes an m6A demethylase implicates m6A as an important regulator of physiological processes. Here, we present a method for transcriptome-wide m6A localization, which combines m6A-specific methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-Seq). We use this method to identify mRNAs of 7,676 mammalian genes that contain m6A, indicating that m6A is a common base modification of mRNA. The m6A modification exhibits tissue-specific regulation and is markedly increased throughout brain development. We find that m6A sites are enriched near stop codons and in 3 UTRs, and we uncover an association between m6A residues and microRNA-binding sites within 3 UTRs. These findings provide a resource for identifying transcripts that are substrates for adenosine methylation and reveal insights into the epigenetic regulation of the mammalian transcriptome.”
Further, “We found that m6A is present in many mRNAs encoded by genes linked to human diseases, including cancer as well as several brain disorders, such as autism, Alzheimer’s disease, and schizophrenia,” says the study’s lead investigator, Dr. Kate Meyer, a postdoctoral researcher in Dr. Jaffrey’s laboratory(ref).”
6. MicroRNAs can play important roles in tumorigenesis.
The June 2012 publication CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer (Ferreira et. al.) reports “Much effort in cancer research has focused on the tiny part of our genome that codes for mRNA. However, it has recently been recognized that microRNAs also contribute decisively to tumorigenesis. Studies have also shown that epigenetic silencing by CpG island hypermethylation of microRNAs with tumor suppressor activities is a common feature of human cancer. The importance of other classes of non-coding RNAs, such as long intergenic ncRNAs (lincRNAs) and transcribed ultraconserved regions (T-UCRs) as altered elements in neoplasia, is also gaining recognition. Thus, we wondered whether there were other ncRNAs undergoing CpG island hypermethylation-associated inactivation in cancer cells”
“Cellular aging is linked to deficiencies in efficient repair of DNA double strand breaks and authentic genome maintenance at the chromatin level. Aging poses a significant threat to adult stem cell function by triggering persistent DNA damage and ultimately cellular senescence.” Further, “65% of naturally occurring repairable DNA damage in self-renewing adult stem cells occurs within transposable elements” of Alu RNA/DNA.
The 2011 publication Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal (Lunyak et. al.) reports “Cellular aging is linked to deficiencies in efficient repair of DNA double strand breaks and authentic genome maintenance at the chromatin level. Aging poses a significant threat to adult stem cell function by triggering persistent DNA damage and ultimately cellular senescence. Senescence is often considered to be an irreversible process. Moreover, critical genomic regions engaged in persistent DNA damage accumulation are unknown. Here we report that 65% of naturally occurring repairable DNA damage in self-renewing adult stem cells occurs within transposable elements. Upregulation of Alu retrotransposon transcription upon ex vivo aging causes nuclear cytotoxicity associated with the formation of persistent DNA damage foci and loss of efficient DNA repair in pericentric chromatin. This occurs due to a failure to recruit of condensin I and cohesin complexes. Our results demonstrate that the cytotoxicity of induced Alu repeats is functionally relevant for the human adult stem cell aging. Stable suppression of Alu transcription can reverse the senescent phenotype, reinstating the cells’ self-renewing properties and increasing their plasticity by altering so-called “master” pluripotency regulators.”
According to Wikipedia : “An Alu element is a short stretch of DNA originally characterized by the action of the Alu restrictionendonuclease[1]. Alu elements of different kinds occur in large numbers in primate genomes. In fact, Alu elements are the most abundant Transposable elements in the human genome.”
“Pairs of human chromosomes. 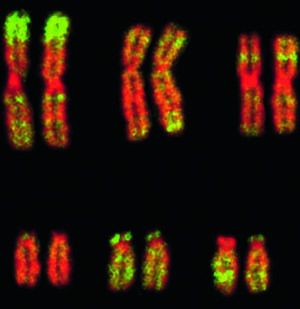 Highlighted in green is alu, a ‘junk’ DNA sequence that distinguishes primates from other mammals.” Image and title source.
Highlighted in green is alu, a ‘junk’ DNA sequence that distinguishes primates from other mammals.” Image and title source.
Specific interventions involving removal of damaged specific segments of RNA, formerly thought to be “junk RNA,” can reverse adult stem cell senescence. Working with specific segments of RNA can add to the traditional epigenetic interventions that mainly have related to DNA methylation and histone acetylation. Specifically, by modifying a Lentivirus genome to express GFP and sh-RNA against Alu transcript, it is possible to knock down the generic SINE/Alu transcript in senescent adult stem cells, reversing senescence markers, rejuvenating the cells, and restoring their lost differentiation capability.
Also from the 2012 publication Protein interactions with piALU RNA indicates putative participation of retroRNA in the cell cycle, DNA repair and chromatin assembly (Blackwell, Luynac et al): “Recently, we have reported that the majority of the repairable DNA damage sites in self-renewing human adult stem cells is associated with the retrotransposal portion of the genome, in particular, Alu retrotransposons.7 Moreover, it has been shown that the upregulation of transcriptional activity from Alu can be triggered by genotoxic stress-induced damage30 as well as upon in-vivo and ex-vivo aging in human retinal pigment epithelium (RPE)31 and human adult stem cells,7 respectively. Intriguingly, evidence indicates that cellular cytotoxicity resulting from increased accumulation of Alu RNA is directly linked to Dicer-1 deficiency,4 suggesting that such cytotoxicity may not necessarily be due to an increase in transcriptional activity, but rather the absence of concomitant transcript processing. Our recently published data indicate that the accumulation of unprocessed Alu transcripts triggers chromatin deterioration, loss of DNA repair in pericentromeric areas eliciting the persistent DNA damage response and, ultimately, cellular senescence.7 — Further, we have shown that human adult stem cells stably expressing an shRNA against Alu transcripts, sh-132Alu, override cellular senescence and reinstate their DNA repair capacity,7 suggesting that RNAi machinery is involved in these events. This published data also implies two equally possible mechanisms through which an shRNA against Alu might mediate the observed function: (1) through the post transcriptional gene silencing (PTGS) Dicer-dependent pathway via the cytoplasmic degradation of Alu RNA or (2) through facilitating transcriptional silencing by recruiting either the Dicer-dependent or Piwidependent arms of the RNAi pathway to act directly on the chromatin as shown in Figure 1A. Both of these pathways are plausible and either could depend on the assembly of single or multiple protein complexes capable of cross-talk with DNAdamage-sensing/repair and centromeric maintenance pathways.”
Graph source: (2011)Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal Lunyak et. al. Cell Cycle. 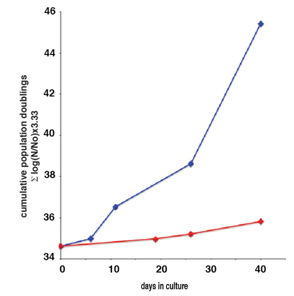 The red line is for senescent hADCs and the blue line shows reinstated proliferation of prior senescent cells.
The red line is for senescent hADCs and the blue line shows reinstated proliferation of prior senescent cells.
Similarly, β-galactoside staining and PCR analyses show that the treated adult stem cells exhibit younger phenotypes.
Wrapping it up
Taking the research all together, it appears that what is happening is adult stem cell rejuvenation via correction of segments of adult stem cell DNA responsible for repair that have themselves become damaged. I think this is a very fundamental line of research. For one matter, it clearly demonstrates epigenetic reversibility of stem cell senescence, a matter some researchers have thought to be impossible. In terms of the Stem Cycle supply chain, the research also suggests that in may not be necessary to revert cells to iPSC status to close the stem cell supply chain loop. It may be sufficient to start with pluripotent adult stem cells, probably adipose-derived ones which are common and relatively easy to collect(ref). These are easier to revert to iPSC status than somatic cells, but such full reversion might not be necessary or desirable.
Of course, further research will be required to develop the in-vivo and eventually human counterparts of this research. How do the formerly senescent adult stem cells expressing an shRNA against Alu transcripts compare with iPSCs? What happens when rejuvinated formerly senescent hADS cells are inserted back into the animal they came from? What will be the consequence on health and longevity? Will there be glitches in the process such as tumorgenesis? Can the entire process of epigenetic reversion of senescent stem cells be safely induced in-vivo? Exactly how? Can this be the basis of an approach to restoring health and youth to humans?
There is still a way to go but the path itself is exciting.